By Dr Fiona McMillan
Researchers at the University of Queensland Diamantina Institute have published a unique method that enables them to use real time, 3 Dimensional imaging to visualise the growth of melanoma tumours. Associate Professor Nikolas Haass, who lead the development of the new method, believes this new approach will help advance melanoma research around the world by enabling scientists to see not only which areas of a tumour respond to a drug, but when.
Haass’ research has revealed that tumour cells may or may not respond to a drug depending on where it is in its growth cycle. This means timing is everything.
“It’s incredibly important to follow the cell cycle in real time,” says Haass.
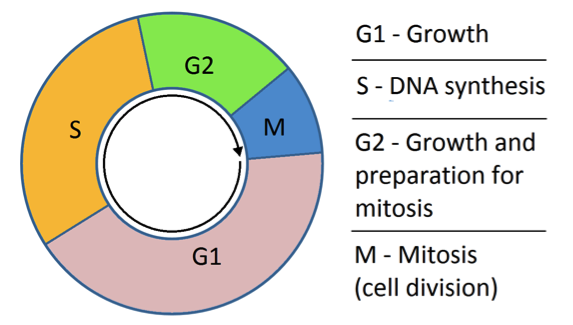
The cell cycle refers to the progression of stages where a cell grows and prepares to divide. These are G1 (for cell growth), S (when the DNA is being replicated), G2 (more growth in preparation for cell division); and Mitosis (when the cell nucleus, including the genetic material, divides). This ultimately leads to cytokinesis (when the entire cell divides into two cells).
While there are some physical changes that occur during the cell cycle, these alone are not enough to accurately pin down which particular phase the cell is in.
The new technique involves modifying certain genes in melanoma cells so that the cells change colour as the cell cycle progresses. The gene that makes the protein CDT1 gets turned on only in G1. The gene that makes the protein called Geminin is only turned on in G2. To take advantage of this, Associate Professor Haass teamed up with researchers at RIKEN in Japan, led by Atsushi Miyawaki.
Miyawaki and his colleagues modified the G1 phase gene (CDT1), so that it would have a fluorescent molecule attached to it. Then, Haass and his team of researchers at UQDI modified melanoma cells so they would now have the new ‘tagged’ version of CDT1. This way, as soon as the gene is turned on and the protein is made, the fluorescent molecule gives off red light until the moment the protein is broken down at the end of G1. In other words, cells fluorescing red are in G1 phase. The same approach was then taken with the G2 phase protein Geminin. But this time, the gene was modified so that it carried a green fluorescent molecule so that cells in G2 phase give off green light.
Of course, the cell cycle goes through more than just these two phases, so what about the others? When the researchers combine the colour coding approach with an analysis of the physical appearance of the cell, it becomes possible to identify where any cell is in its cycle at any point in time.
Crucially, Haass and his colleagues are able to use this method to look at how different tumour cells cycle, or stay dormant, in a 3D model of melanoma.
Traditionally, technology limitations have meant that tumour biology is studied in 2 dimensions: the tumour cells are spread out side by side in layers on a culture. With this 2D approach, each cell has the same access to nutrients. But this isn’t an accurate representation of a real tumour in the body, where the tumour structure, as well as the way the cells interact with each other and the local environment, are much more complex. To overcome the limitations of 2D, Haass developed 3D tumour spheroids.
“We do almost everything in 3D,” he says. “It allows us to mimic the tumour architecture and the microenvironment. We see a number of things in 3D culture that you just don’t see in 2D, and this much more closely approximates what happens with tumours in real life.”
“We can embed these 3D spheroids into a collagen matrix so we can study both the tumour growth, and its ability to invade surrounding tissue at the same time.”
With the colour changing method, this means that he and his colleagues can analyse 3D melanoma tumours as time progresses.
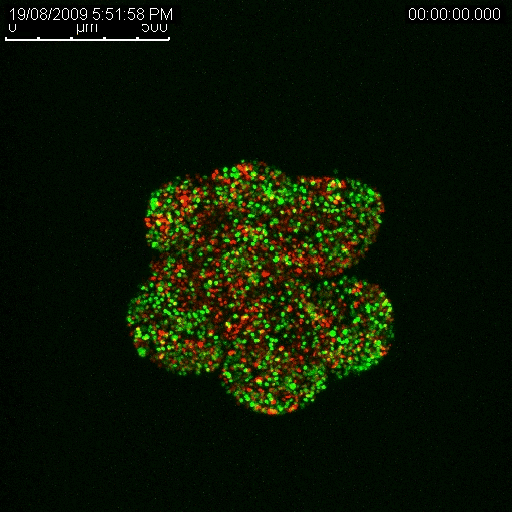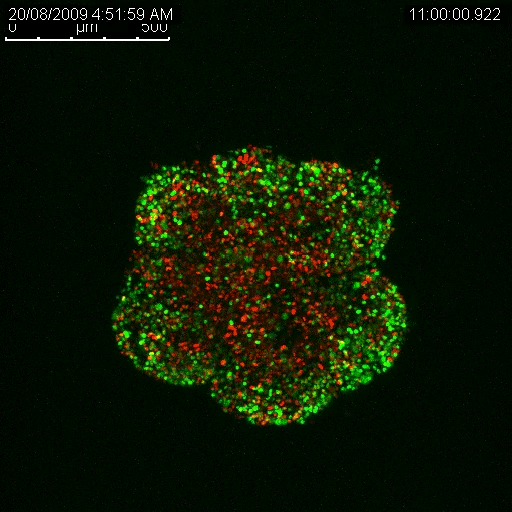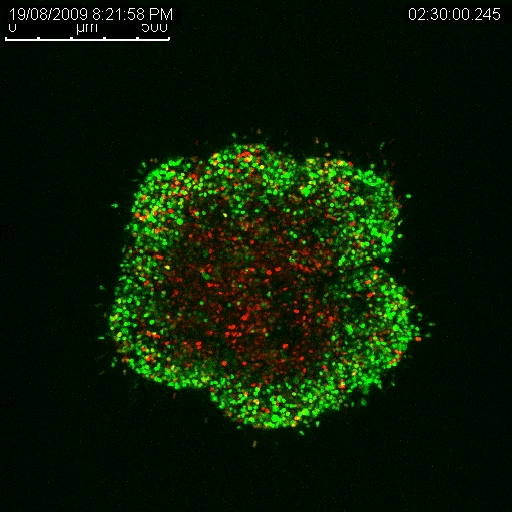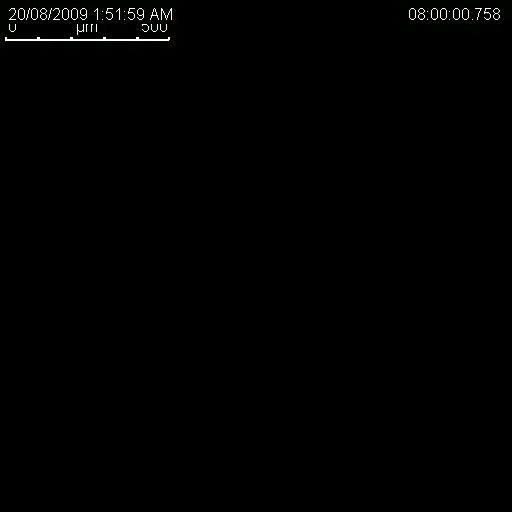
Haass’ research has enabled him to determine that there are separate populations of cancerous cells in melanoma tumours, and that some are growing at different rates and some are dormant (Haass et al. 2014, Pigment Cell Melanoma Res.). The dormant ones are the big problem when it comes to drug resistance, he explains. They are not as responsive to treatment as the actively growing cells precisely because many anti-cancer treatments target and disrupt growth functions in a cell. If a cell isn’t growing, the treatment has little to no effect.
Haass is also using this method to investigate the line of separation between the growing cells and the cells that remain dormant. By finding out what’s going on at that boundary, he hopes to learn what enables some cancer cells to go to sleep and resist treatment, only to awaken at a later time. Such knowledge could have big implications for improving the effectiveness of many anti-cancer therapeutics. The key will be finding a way to safely wake up sleeping tumour cells just enough to make them vulnerable and kill them.
The video article, along with a full list of collaborators, can be found here:
Beaumont, K. A., Anfosso, A., Ahmed, F., Weninger, W., Haass, N. K. Imaging- and Flow Cytometry-based Analysis of Cell Position and the Cell Cycle in 3D Melanoma Spheroids. J. Vis. Exp. (106), e53486, doi:10.3791/53486 (2015).
MEDIA: Kate Templeman on 0409 916 801 or k.templeman@uq.edu.au